| Bos taurus Gene: CLDN3 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Summary | |||||||||||||||
| InnateDB Gene | IDBG-648721.3 | ||||||||||||||
| Last Modified | 2014-10-13 [Report errors or provide feedback] | ||||||||||||||
| Gene Symbol | CLDN3 | ||||||||||||||
| Gene Name | Claudin-3 | ||||||||||||||
| Synonyms | |||||||||||||||
| Species | Bos taurus | ||||||||||||||
| Ensembl Gene | ENSBTAG00000040432 | ||||||||||||||
| Encoded Proteins |
Claudin-3
|
||||||||||||||
| Protein Structure |

|
||||||||||||||
| Useful resources | Stemformatics EHFPI ImmGen | ||||||||||||||
| Entrez Gene | |||||||||||||||
| Summary |
This gene does not have any Entrez summary - the following is the summary from its human ortholog ENSG00000165215:
Tight junctions represent one mode of cell-to-cell adhesion in epithelial or endothelial cell sheets, forming continuous seals around cells and serving as a physical barrier to prevent solutes and water from passing freely through the paracellular space. These junctions are comprised of sets of continuous networking strands in the outwardly facing cytoplasmic leaflet, with complementary grooves in the inwardly facing extracytoplasmic leaflet. The protein encoded by this intronless gene, a member of the claudin family, is an integral membrane protein and a component of tight junction strands. It is also a low-affinity receptor for Clostridium perfringens enterotoxin, and shares aa sequence similarity with a putative apoptosis-related protein found in rat. [provided by RefSeq, Jul 2008] |
||||||||||||||
| Gene Information | |||||||||||||||
| Type | Protein coding | ||||||||||||||
| Genomic Location | Chromosome 25:34018456-34019669 | ||||||||||||||
| Strand | Forward strand | ||||||||||||||
| Band | |||||||||||||||
| Transcripts |
|
||||||||||||||
| Interactions | |||||||||||||||
| Number of Interactions |
This gene and/or its encoded proteins are associated with 0 experimentally validated interaction(s) in this database.
They are also associated with 7 interaction(s) predicted by orthology.
|
||||||||||||||
| Gene Ontology | |||||||||||||||
Molecular Function |
|
||||||||||||||
| Biological Process |
|
||||||||||||||
| Cellular Component |
|
||||||||||||||
| Orthologs | |||||||||||||||
|
Species
Homo sapiens
Mus musculus
|
Gene ID
Gene Order
|
||||||||||||||
| Pathways | |||||||||||||||
| NETPATH | |||||||||||||||
| REACTOME |
Cell junction organization pathway
Cell-Cell communication pathway
Cell-cell junction organization pathway
Tight junction interactions pathway
|
||||||||||||||
| KEGG | |||||||||||||||
| INOH | |||||||||||||||
| PID NCI | |||||||||||||||
| Pathway Predictions based on Human Orthology Data | |||||||||||||||
| NETPATH | |||||||||||||||
| REACTOME |
Tight junction interactions pathway
Cell-Cell communication pathway
Cell junction organization pathway
Cell-cell junction organization pathway
|
||||||||||||||
| KEGG |
Cell adhesion molecules (CAMs) pathway
Leukocyte transendothelial migration pathway
Tight junction pathway
Hepatitis C pathway
Tight junction pathway
Cell adhesion molecules (CAMs) pathway
Leukocyte transendothelial migration pathway
Hepatitis C pathway
|
||||||||||||||
| INOH | |||||||||||||||
| PID NCI | |||||||||||||||
| Cross-References | |||||||||||||||
| SwissProt | Q765N9 | ||||||||||||||
| TrEMBL | Q765P0 | ||||||||||||||
| UniProt Splice Variant | |||||||||||||||
| Entrez Gene | 404153 | ||||||||||||||
| UniGene | Bt.6074 | ||||||||||||||
| RefSeq | NM_205801 | ||||||||||||||
| HUGO | |||||||||||||||
| OMIM | |||||||||||||||
| CCDS | |||||||||||||||
| HPRD | |||||||||||||||
| IMGT | |||||||||||||||
| EMBL | AB115780 AB115781 BC102398 | ||||||||||||||
| GenPept | AAI02399 BAD01112 BAD01113 | ||||||||||||||
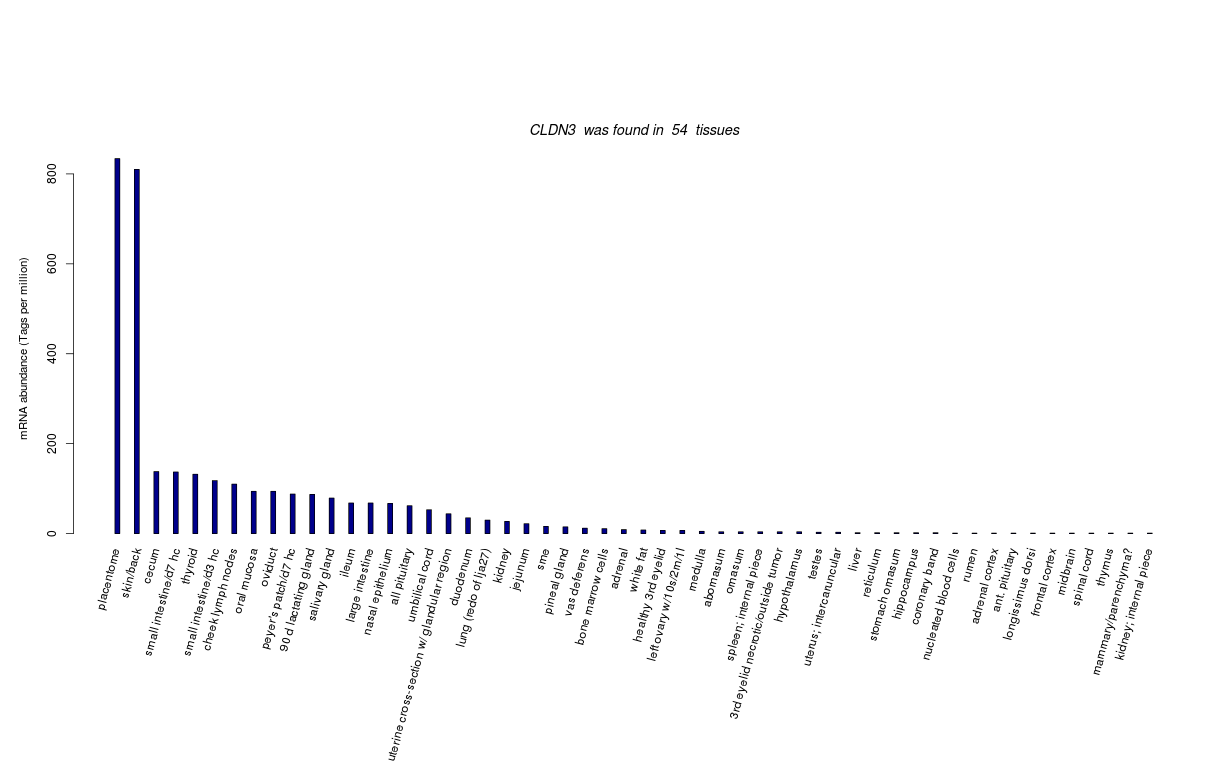
| RNA Seq Atlas | 404153 | ||||||||||||||
| Transcript Frequencies | |||||||||||||||
| Tag Count based mRNA-Abundances across 87 different Tissues (TPM).
Based on Data from Bovine Gene Atlas |
(Move your mouse over the image to view a more detailed version) |
||||||||||||||